Chapter 17 Using Dictionary on Biological Data
We will practice using dictionary by solving a set of problems. First, we will solve them using the Python rules covered so far. You will see that extending this solution poses a challenge. We will introduce the idea of dictionary to overcome that challenge.
17.1 Problem - Counting of nucleotides
The simplest task in genetics is to count the number of nucleotides and determine their percentages. That number is used in many experiments and analyses. Can you tell me how many A, T, G, Cs are present in the following part of SARS-CoV-2 genome?
ATTAAAGGTTTATACCTTCCCAGGTAACAAACCAACCAACTTTCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTTAAAATCTGTGTGGCTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATTAATAACTAATTACTGTCGTTGACAGGACACGAGTAACTCGTCTATCTTCTGCAGGCTGCTTACGGTTTCGTCCGTGTTGCAGCCGATCATCAGCACATCTAGGTTTCGTCCGGGTGTGACCGAAAGGTAAGATGGAGAGCCTTGTCCCTGGTTTCAACGAGAAAACACACGTCCAACTCAGTTTGCCTGTTTTACAGGTTCGCGACGTGCTCGTACGTGGCTTTGGAGACTCCGTGGAGGAGGTCTTATCAGAGGCACGTCAACATCTTAAAGATGGCACTTGTGGCTTAGTAGAAGTTGAAAAAGGCGTTTTGCCTCAACTTGAACAGCCCTATGTGTTCATCAAACGTTCGGATGCTCGAACTGCACCTCATGGTCATGTTATGGTTGA17.1.1 STEP 1 - Simplify the Problem
In our simplification, we will use x = ATTAAA as the sequence and count only the number of As.
You see it is a vastly simpler problem. The sequence has only 6 nucleotides, and they can be accessed as x[0], x[1], x[2], x[3], x[4], x[5].
So the code will look like -
x="ATTAAA"
count_A = 0
if(x[0] =='A'):
count_A = count_A + 1
if(x[1] =='A'):
count_A = count_A + 1
if(x[2] =='A'):
count_A = count_A + 1
if(x[3] =='A'):
count_A = count_A + 1
if(x[4] =='A'):
count_A = count_A + 1
if(x[5] =='A'):
count_A = count_A + 1
print(count_A)Let me explain what is going on. In line 1, Python creates a box and puts the value “ATTAAA.”
In line 2, it creates a box (variable) count_A and put the value 0.
In line 4, Python checks if x[0] is equal to ‘A.’ If so, count_A is incremented by 1 in the following line.
We do the same check 5 more times for other letters of x. Then we print the count_A at the end.
17.1.2 STEP 2 - Replace with Loop
If we like to expand this code to 100 letters, that will require a lot of typing. Can we use a loop instead?
x="ATTAAA"
count_A = 0
for i in [0, 1, 2, 3, 4, 5]:
if(x[i] =='A'):
count_A = count_A + 1
print(count_A)Above code creates a “box” i in line 4 and stores the value 0. Then it runs the following two lines with i=0, and that is equivalent to lines 4 and 5 of the earlier code without loop.
Then the same loop runs for i = 1, 2 and so on until 5. Thus this code is identical to the previous one without loop.
In the next step, we replace the list in the for loop` with range. We notice that [0, 1, 2, 3, 4, 5] is just range(6), but where does 6 comes from? It is the length of x. So, we write -
x="ATTAAA"
count_A = 0
L=len(x)
for i in range(L):
if(x[i] =='A'):
count_A = count_A + 1
print(count_A)This takes care of input sequence of arbitrary length to count As, but how about Cs, Gs and Ts?
You will notice that if we replace ‘A’ by ‘C’ in the code from line 2 onward, the program will count Cs.
To count all “A,” “C,” “G,” “T,” we write -
x="ATTAAA"
count_A = 0
count_C = 0
count_T = 0
count_G = 0
L=len(x)
for i in range(L):
if(x[i] =='A'):
count_A = count_A + 1
if(x[i] == 'C'):
count_C = count_C + 1
if(x[i] == 'T'):
count_T = count_T +1
if(x[i] =='G'):
count_G = count_G + 1
print(count_A, count_T, count_C, count_G)17.2 Problem - Counting of nucleotide pairs and triplets
We are going to extend the above code to counting pairs (“AA,” “AT,” etc.) and triplets (“AAA,” “AAT,” etc.). You will find that it gets challenging. For pairs, you will need to keep track of 16 count variables (count_AA, count_AC, etc.). The number will increase to 64 for triplets. Imagine if you had to count all 30-letter words, which is a common question in genomics.
17.2.1 Introducing Dictionary
Instead of a list with indices 0, 1, 2, .., we will have a data structure with words as indices. Let me show an example.
x={}
x['a']=1
x['b']=2
print(x)In the first line, we are creating an empty “dictionary” x. In the second line, we are inserting the value 1 in position a of x. In the second line, we are inserting the value 2 in position b of x.
Here “a” and “b” called keys, and 1 and 2 are the corresponding values.
Another way to create a dictionary is this -
x={'a': 1, 'b' : 2, 'c' : 4}
print(x)Note the difference with a list - x = [1, 2, 4]
The list x has values 1, 2, 4 respectively in positions x[0], x[1], x[2]
The dictionary x has 1, 2, 4 in the positions x[‘a’], x[‘b’], x[‘c’].
Also, the positions in the dictionary are not ordered. That means if you run print(x), it may print the keys and values in any order.
17.2.1.1 Exercise
Can you create a dictionary with the names and capitals of four countries?
17.2.1.2 Solution
x = {'Japan' : 'Tokyo', 'China' : 'Beijing', 'Italy': 'Rome', 'Russia': 'Moscow'}
print(x)
print(x['Japan')With a dictionary, you can ask questions like “What is the capital of Japan?” and get the answer quickly.
17.2.2 Solving Previous Counting Problem Using Dictionary
Next, we will see how a dictionary can be used to simplify yesterday’s solution for counting nucleotides. Let me first paste the code from yesterday.
Instead of having four variables count_A, count_C and so on, we will have a dictionary.
x="ATTAAA"
count={'A': 0, 'C': 0, 'G': 0, 'T': 0 }
L=len(x)
for i in range(L):
count[x[i]] = count[x[i]]+1
print(count)Let me explain, how this code is equivalent to the previous one.
Previously, you had -
count_A = 0
count_C = 0
count_T = 0
count_G = 0That is replaced by -
count={'A': 0, 'C': 0, 'G': 0, 'T': 0 }Instead of variables called count_A etc., we have count['A']. The dictionary condenses 4 different variables into one.
Understanding the following line is hard. Let us do that by expanding the loop.
for i in range(L):
count[x[i]] = count[x[i]]+1By substituting i with 0, 1, 2, …5, the loop becomes -
count[x[0]] = count[x[0]]+1
count[x[1]] = count[x[1]]+1
count[x[2]] = count[x[2]]+1
count[x[3]] = count[x[3]]+1
count[x[4]] = count[x[4]]+1
count[x[5]] = count[x[5]]+1By substituting x[0] to x[5], the code becomes -
count['A'] = count['A']+1
count['T'] = count['T']+1
count['T'] = count['T']+1
count['A'] = count['A']+1
count['A'] = count['A']+1
count['A'] = count['A']+117.2.3 Solving The Problem of Counting Pairs
Let us now try to use this new tool to solve the problem of counting pairs. Here we are asked to count all overlapping pairs.
Example:
Input - “ATTAAA,”
Output - “AA”/2, “AT”/1, “TT”/1, “TA”/1.
First you create a dictionary with all possible pairs.
x="ATTAAA"
count={'AA': 0, 'AC': 0, 'AG': 0, 'AT': 0, 'CA': 0, 'CC': 0, 'CG': 0, 'CT': 0, 'GA': 0, 'GC': 0, 'GG': 0, 'GT': 0, 'TA': 0, 'TC': 0, 'TG': 0, 'TT': 0 }Next we will retrace the steps of the previous dictionary example, but in reverse. In the expanded form, the counting part of the code is -
count["AT"]=count["AT"]+1
count["TT"]=count["TT"]+1
count["TA"]=count["TA"]+1
count["AA"]=count["AA"]+1
count["AA"]=count["AA"]+1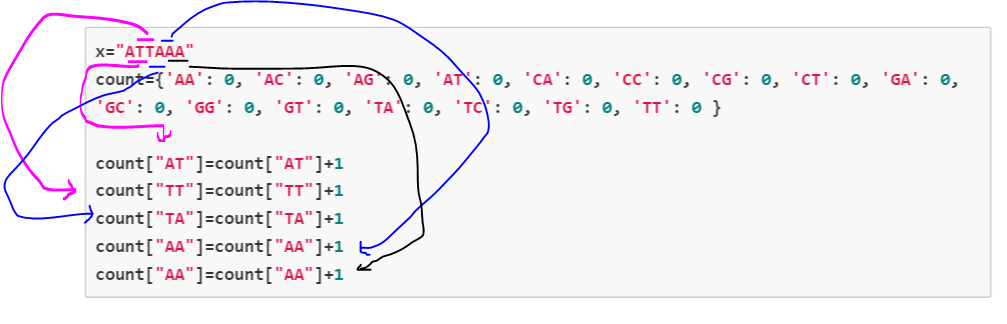
Image
Where do those words “AT,” “TT” etc. come from? They are actually parts of the original word. Therefore, you can write the above code as -
count[x[0:2]]=count[x[0:2]]+1
count[x[1:3]]=count[x[1:3]]+1
count[x[2:4]]=count[x[2:4]]+1
count[x[3:5]]=count[x[3:5]]+1
count[x[4:6]]=count[x[4:6]]+1Let us replace that with a code -
for i in range(len(x)-1):
count[x[i:i+2]]=count[x[i:i+2]]+1Therefore, the entire code becomes -
x="ATTAAA"
count={'AA': 0, 'AC': 0, 'AG': 0, 'AT': 0, 'CA': 0, 'CC': 0, 'CG': 0, 'CT': 0, 'GA': 0, 'GC': 0, 'GG': 0, 'GT': 0, 'TA': 0, 'TC': 0, 'TG': 0, 'TT': 0 }
for i in range(len(x)-1):
count[x[i:i+2]]=count[x[i:i+2]]+1
print(count)17.3 Exercise
Check whether you can extend the above code to count the number of triplet words?